The BRIGHT Biofoundry
Where practical knowledge arises in the melting pot between wet biology, dry biology, big data, machine learning and artificial intelligence.
The BRIGHT Biofoundry is a dynamic, technology-driven research hub dedicated to accelerating innovation in biotechnology, with a particular focus on the design and development of microbial cell factories and novel food systems. It seamlessly integrates key enabling technologies within a multidisciplinary ecosystem, combining computational and molecular biology to transform large-scale biological data into actionable insights — all while adhering to the FAIR (Findable, Accessible, Interoperable, Reusable) data principles. Operating from TRL0 to TRL6 — from early-stage ideas to the creation of spin-offs — and supporting bioprocess development from nanoliter to 100-liter scale, the BRIGHT Biofoundry drives the entire innovation pipeline. To ensure that projects are both economically viable and impactful in advancing biosustainability, it maintains strong collaborations with the Sustainable Innovation Office and the Commercialization and IP Support team.
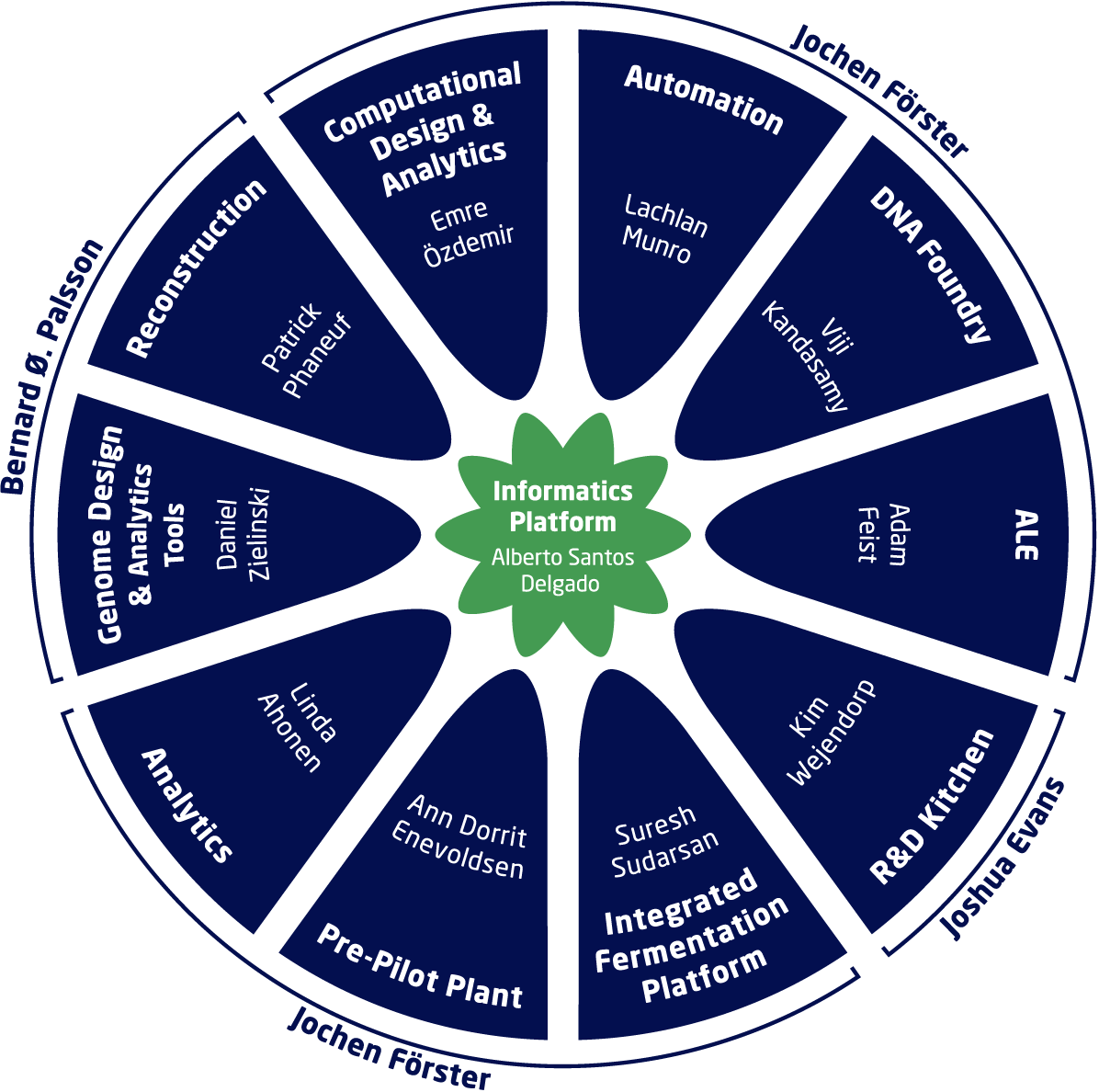
The BRIGHT Biofoundry comprises 11 specialized teams, each contributing unique capabilities to the research pipeline:
- Automation: Streamlining laboratory processes to enhance throughput and reproducibility.
- DNA Foundry: Designing and assembling complex genetic constructs and microbial cell factories with high precision.
- Adaptive Laboratory Evolution: Engineering microbial traits through evolutionary strategies.
- R&D Kitchen: Rapid prototyping and experimental development of novel food systems.
- Integrated Fermentation Platform: Scaling microbial processes under controlled bioprocessing conditions.
- Pre-Pilot Plant: Bridging laboratory and industrial-scale production for process validation.
- Analytics: Providing high-resolution, quantitative analysis of biological samples.
- Genome Design & Analytics Tools: Developing rational genome engineering and data interpretation tools.
- Reconstruction: Systematically modelling and simulating biological networks using pan omics analysis.
- Computational Design & Analytics: Utilizing AI and data science to optimize biological design and interpret biological data.
- Informatics Platform: Delivering FAIR-compliant data management infrastructure, shared ontology, and centralized interfaces to support data-driven project execution.
The Biofoundry supports three types of projects:
- Large Biofoundry Projects The goal of Large Biofoundry Projects is to bridge the technological "valley of death" and advance selected research in developing microbial cell factories for chemical or protein production. These projects aim to enhance research value and commercial potential, such as spin-offs or licensing deals. The Biofoundry adds resources to accelerate progress to a first prototype. Projects are finalized in the Pre-Pilot Plant for optimization and scale-up, enabling spin-outs or licensing opportunities.
2. Biofoundry Support Projects The primary aim is to conduct tasks requested by research groups of our centre or our sister departments at DTU. We also collaborate with external partners, including other universities and industry. Tasks may include omics projects, automation support, and adaptive laboratory evolution projects, etc.
3. Biofoundry Technology Development Projects The focus is on upgrading our technology platform or adding novel technologies that will enable optimized cell factory development.
Contact
Jochen Förster Professor, Director BRIGHT Biofoundry jfor@dtu.dk