A new cutting edge biological sensor have led to the discovery of 25 transporter proteins in bacteria. Transporters control bacterial communication and their uptake of different compounds such as drugs and vitamins. This discovery points towards new antibiotics and more efficient biological production of vitamins.
Researchers from The Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain) at Technical University of Denmark, and the startup company Biosyntia has developed a molecular sensor system – a biosensor – that reveals transporter proteins in the bacteria’s membrane.
Transporter proteins are embedded in the cell membrane and actively transport certain molecules in and out of the organism. This group of proteins is extremely interesting to investigate in order to understand how, for instance, intestinal bacteria exchange nutrients and hence affect our gut health.
Finding – and targeting – bacterial transporter proteins
Using this new biosensor, the researchers found 25 novel bacterial proteins transporting vitamin B1 (thiamine). Some pathogenic microorganisms, so-called auxotrophs, cannot produce important vitamins themselves and depend on uptake from the surrounding environment. Hence, transporters of essential metabolites constitute potent drug targets.
The bacterium Helicobacter pylori, which causes gastric ulcer and gastric cancer, can't produce B1 itself and dependents on uptake from the external environment for growth. The research led to the identification of the vitamin uptake mechanism, which was unknown before the study.
"With this knowledge, you will most likely be able to develop drugs that can block transporter proteins and, hence, kill pathogenic bacteria such as H. pylori," says Hans Genee, main author and co-founder of the biotech company Biosyntia.
"When we understand the interactions between bacteria, we can begin to cure intestinal and infectious diseases with new types of antibiotics with different molecular targets than conventional antibiotics"
Professor Morten Sommer
The comprehensive study has now been published in the prestigious journal Nature Chemical Biology.
Novel vitamin transporters from gut and soil bacteria
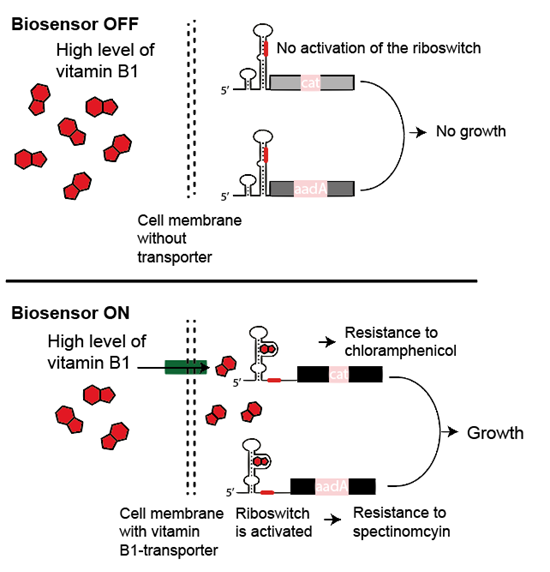
The biosensor works by a so-called riboswitch, which is a piece of RNA – the "intermediate" between genes and proteins – that can turn on and off certain genes.
The researchers' artificial riboswitch was designed to bind vitamin B1 (thiamine), but only when the concentration of B1 was very high in the cell. The binding of B1 activated two genes conferring resistance to the antibiotics chloramphenicol and spectinomycin.
In a series of experiments, the researchers screened millions of E. coli containing both the riboswitch and a random piece of foreign DNA from intestinal and soil bacteria. This group of bacteria is interesting for human health, but unfortunately most are hard to grow and hence examine experimentally. This new method is therefore key to understanding this novel landscape.
The next step was to grow E. coli cells with vitamin B1, chloramphenicol and spectinomycin. In this way only cells capable of actively taking up B1, could activate the riboswitch, thereby obtaining resistant to the antibiotics. The remainder of the bacteria didn’t grow. In this way the riboswitch revealed that a transporter protein was present in surviving cell colonies.
"By looking at the specific piece of DNA in growing colonies, we could thus identify which gene encoding the transport protein. This has not been done before," says Professor and co-author Morten Sommer from DTU Biosustain.
Understanding ‘bacterial communication’ is important for human health
Until now, transporter proteins have been difficult to identify – despite an intensive research effort. In fact, the function of about 75 percent of all transporter proteins present in microorganisms remain unknown.
"When you look at the DNA of bacteria, many genes appear to encode transporter proteins. But the challenge so far has been to go from gene to showing the function of transporter proteins," says Morten Sommer and continues:
"This discovery is important for understanding how bacteria communicate with each other in, for instance, the gut, where some microbes excrete vitamins and essential amino acids, and other use them. When we understand the interactions, we can begin to cure intestinal and infectious diseases with new types of antibiotics having different molecular targets than conventional antibiotics."
Vitamins can be produced biologically
The biotech company Biosyntia is designing vitamin producing bacteria. Today, the vast majority of vitamins in animal feed and vitamin pills are produced by chemical synthesis, which has a greater negative environmental impact than biological manufacturing.
But in order to design cells – cell factories – that can produce large amounts of vitamins biologically, researchers must engineer the bacteria's genome.
This requires screening of millions of cell variants to find the perfect design. And in this quest, the new biosensor is a revolution, because it can also be modified to isolate cell’s that produce high levels of vitamins.
"Before this method we could screen approximately 100 bacteria per day for their vitamin production in a relatively expensive method called HPLC. Now we can test 100 million bacterial clones in one day in a single petri dish, where only the best cells will show up on the plate. So instead of searching for the needle in a haystack straw by straw, we burn down the haystack," says Hans Genee.
The next step is to find more unknown proteins and examine how they work - and hopefully finding new antibiotics that can block the bacteria's vital transporter proteins.
Other researchers have tried to develop similar systems, but many false-positive has been a major barrier to practical usage. However, by using a dual system in which two biosensors are coupled to two different antibiotic resistance genes, the system significantly reduced false positive growth.
The researchers showed that the biosensor approach works for other substances than B1 and thus can be used to mine other important functions. In order to show the diversity of the biosensor, the researchers constructed a similar system that responded to xanthine – an important intermediate in e.g. caffeine, and used this system to mine xanthine uptake transporters from soil and gut microbiomes.